Tav4SB project
Taverna services for Systems Biology
Contents
Contact
If you have any questions, comments or seek help with our WS or any other resource, please contact us via tav4sb discussion group or directly at tav4sb@mimuw.edu.pl.
Multi-parameter sensitivity analysis (MPSA) of a deterministic and stochastic SBML models.
This is a MPSA experiment for both deterministic and stochastic versions of the enzymatic reaction model (SBML). MPSA is a Monte Carlo filtering method of sensitivity analysis which maps parameters space into acceptable and unacceptable output regions. The method works as follows:
- Select parameters to assess.
- Set parameters range.
- Generate independent samples.
- Calculate samples errors.
- Classify samples as acceptable or unacceptable.
- For each selected parameter compare classified samples sets.
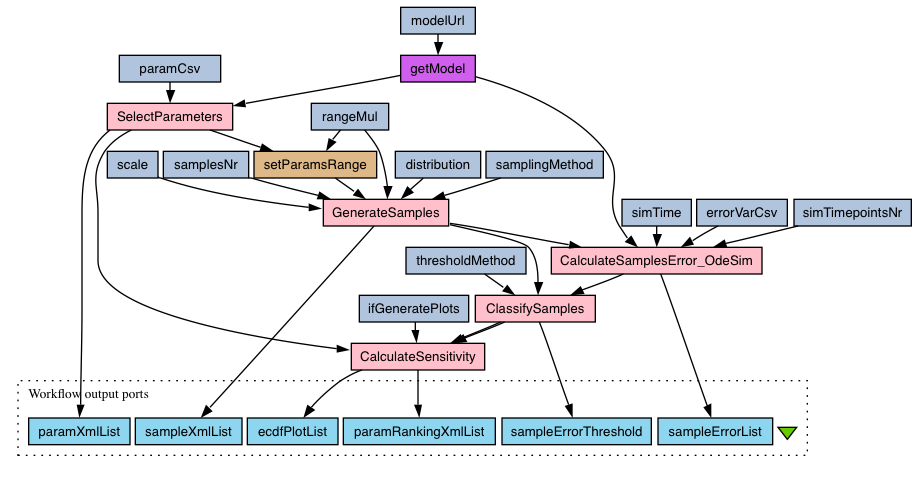 "MPSA of a deterministic SBML model" workflow
"MPSA of a deterministic SBML model" workflow
We focus on kinetic parameters of the two forward reactions, i.e. k1 and k3. As an error we take the mean squared error of the ODE trajectory of product P for the deterministic variant. For the stochastic variant we exploited probabilistic model checking and take the value of the CSL formula, which asks the model about how many times, on average, the enzyme-substrate complex association reaction have to occur before amount of the product reaches 50% of its maximum?. Both error functions are calculated for each parameters sample with respect to results for the original model.
Both MPSA experiments' workflows (t2flow) require the JSBML
library (see JAR libraries in resources).
If you haven't installed it earlier, download and copy the JAR file to
Taverna's
library folder; restart Taverna afterwards.
To run the workflow for deterministic model choose
File > Open workflow location... and copy-paste the
link to the workflow file.
Then, File > Run workflow... and wait for results. Good moment
to grab a snack. This workflow has no inputs - all parameters are embedded
in string constants (e.g. modelUrl, paramCsv,
samplesNr, simTime, errorVarCsv,
thresholdMethod etc). That's where you start if you want to
play around. To run the workflow for stochastic model repeat above steps
using this
link to the workflow file.
For both workflows you can plot the error surface for parameters k1 and k3.
To do that you will need to save MPSA results to file, i.e. from the
Results perspective choose Save all values > Save in single
XML document. Next, open and run
the plotting workflow file.
Taverna Workbench will ask for filling in the input values, so, in the input
values dialog, choose Load previous values and point to previously
saved MPSA values. If you want to play around with plot options use the
string constant processor optionsCSV. Don't forget to check out
the description of plotting options for the mathPlot
WS operation, i.e. the
wrapper docs on
optionList input at myExperiment or
input docs at
BioCatalogue.
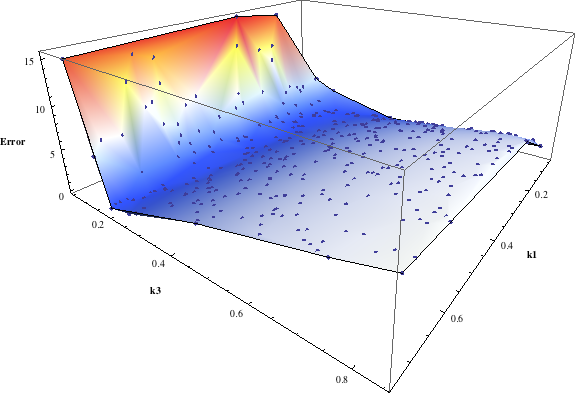
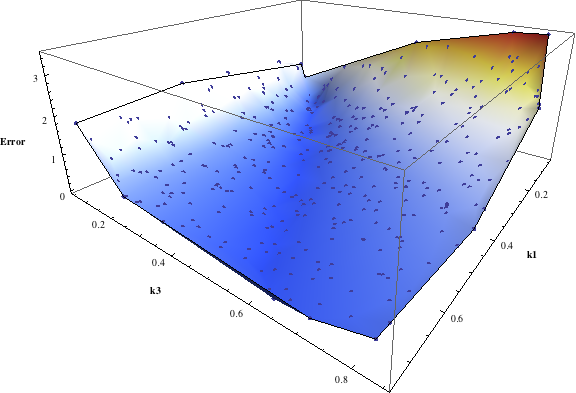
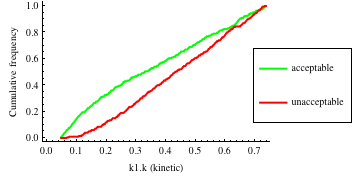
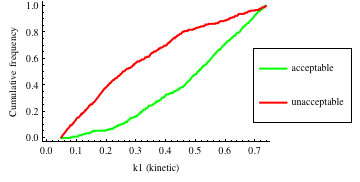
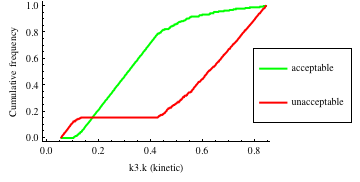
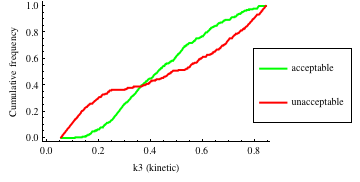
In turn, you will obtain many intermediate outputs of the MPSA procedure, including plots of empirical cumulative distribution functions (ECDF) of acceptable and unacceptable samples, for each of the selected parameters. The final result is <paramRankingXmlList>. It contains two rankings of parameters. Values of rankings are calculated by comparing ECDFs with Kolmogorov-Smirnov test (KS test) and one minus Pearson product-moment correlation coefficient (PMCC). If you also plotted the error surface for both variants, the final set of results should include the following plots and rankings:
| ODEs simulations | PMC | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 |
 |
||||||||||||||||||
 |
 |
||||||||||||||||||
 |
 |
||||||||||||||||||
|
|
In the variant based on ODEs simulations and the error function which measures changes in the product behavior, parameter k3 significantly dominates parameter k1, as far as sensitivity of the system is concerned. This is an expected result: the k3 is a parameter of reaction which is directly responsible for product creation; from the Michaelis-Menten approximation one can expect that, for our model values, variation of the parameter k3 will be more influential, with respect to the product rate, than variation of the parameter k1. Results of the PMC-based variant of the MPSA procedure are opposite: now k1 dominates k3. This may be ascribed to the particular choice of formula which calculates the average number of occurrences of the first reaction. The domination is not as definite as in the first variant of MPSA.
Resources
Taverna 2 workflow files: