Tav4SB project
Taverna services for Systems Biology
Contents
Contact
If you have any questions, comments or seek help with our WS or any other resource, please contact us via tav4sb discussion group or directly at tav4sb@mimuw.edu.pl.
Simulate SBML-derived ODEs
This is a simple ODEs numerical simulation experiment for the enzymatic reaction deterministic model (SBML). ODEs are derived automatically, based on SBML-encoded rate laws for reactions.
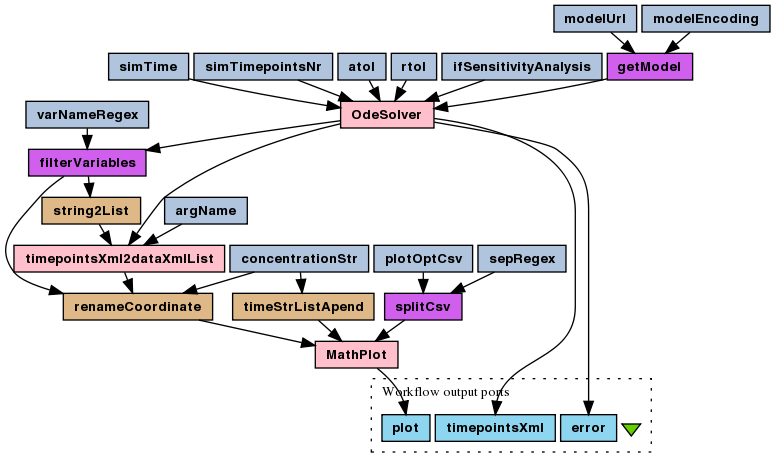
To run this experiment's workflow choose File > Open workflow
location... and copy-paste the following
link to the workflow file.
Then, File > Run workflow... and wait for results.
This workflow has no inputs - all simulation and plot parameters are
embedded in string constants (modelUrl, simTime,
simTimepointsNr and plotMarkersOpt, plus
filterVariables/regex default value). That's what you should
change if you want to play around. In particular, there is only one plotting
option given, which says not to plot data points markers.
Note: For description of plotting options check mathPlot
WS operation
wrapper docs on
optionList input at myExperiment or
input docs at
BioCatalogue.
As a result of running this experiment you should get the
<timepoints> XML element with simulated values of SBML
species variables over 30 seconds, and the following plot of these time
points:
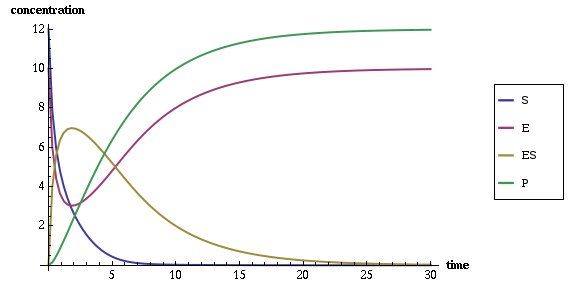
From the plot you can see that the system stabilizes in approximately 25 seconds with the peak activity at 2nd second, when most of the enzymes are at work, being involved with substrate in a complex, which, in turn, converts substrate to the product.
Resources
Taverna 2 workflow files: