A short comparison of network clustering algorithms
Below we compare clusterings retured by affinity propagation, MCODE and MCL.
Our goal here is not in a detailed and comprehensive comparison -- we just want to show that AP should be considered as a valuable tool when searching for modules in weighted networks.
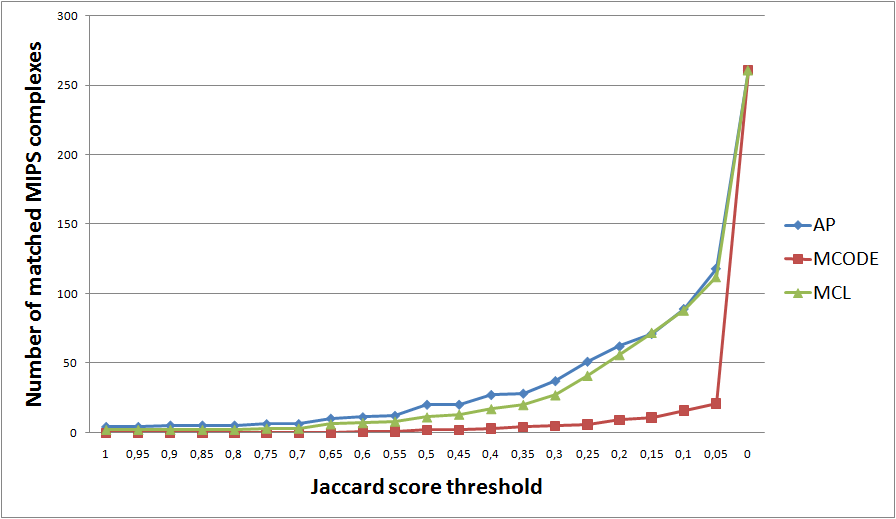
We cluster the PPI network of Saccharomyces cerevisiae (SC). For each MIPS complex we look for the cluster with the best Jaccard score. The graphs below show the number of MIPS complexes (Y axis) for which there exist clusters with Jaccard index greater than or eaqual to the treshold (X axis).
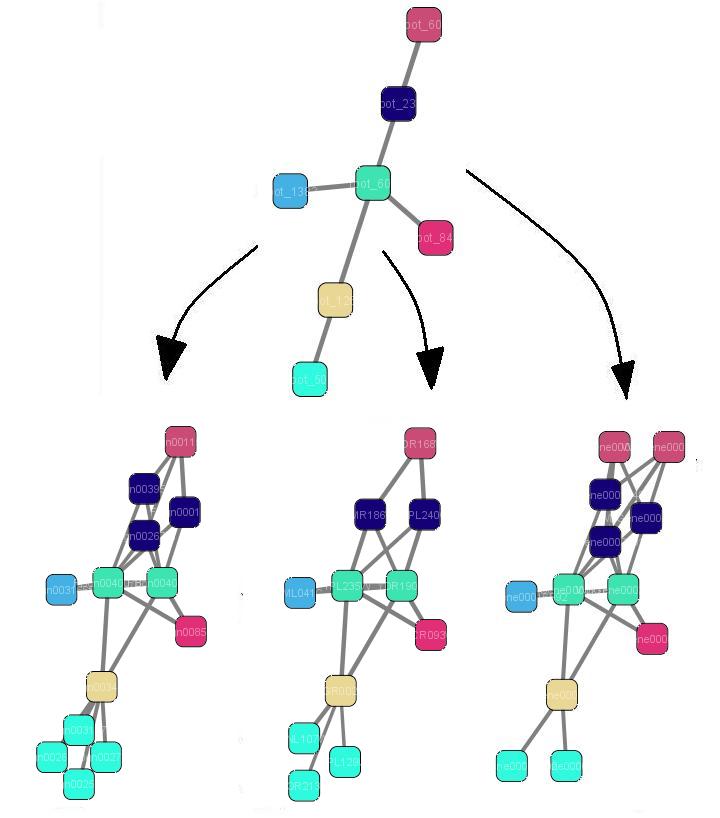
We obtain the clusters in two ways: by directly clustering our default SC network (edge treshold 0.7) or alternatively, by clustering the ancestral network (root) (threshold 0.1, 0.3, or 0.7) and mapping the identified clusters onto the SC network.
-
Clustering of Saccharomyces cerevisiae PPI network (threshold 0.7).

-
Two steps: Clustering of ancestral PPI network and mapping on Saccharomyces cerevisiae (treshold = 0.1).

-
Two steps: Clustering of ancestral PPI network and mapping on Saccharomyces cerevisiae (treshold = 0.3).

-
Two steps: Clustering of ancestral PPI network and mapping on Saccharomyces cerevisiae (treshold = 0.7).