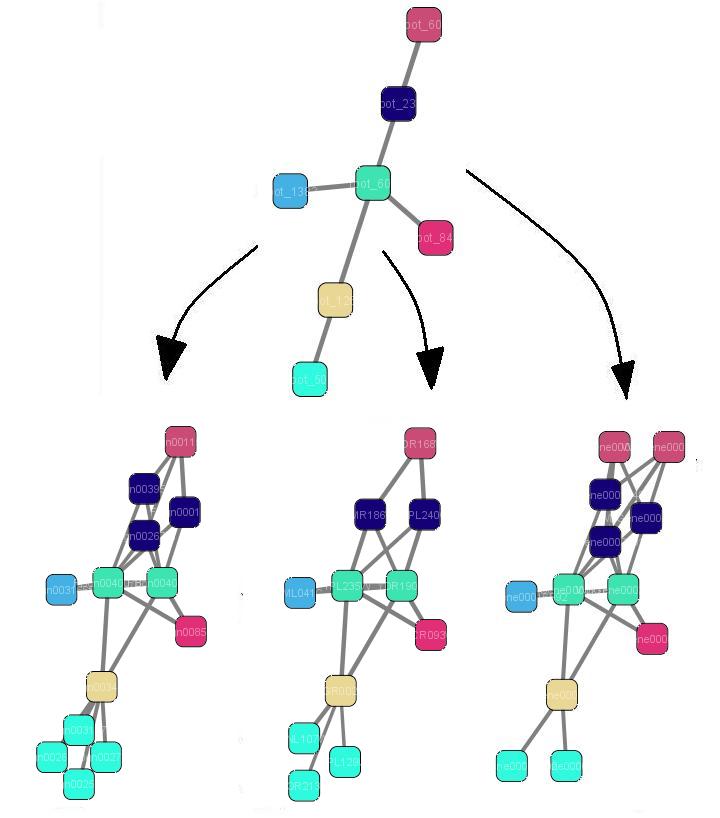
Cytoscape plugins
- NetworkEvolution plugin provides the capability to visualize censecutive evolutionary stages of protein interaction networks based on phylogenetic information.
- APCluster plugin implements a recent affinity propagation algorithm for graph clustering.
All implemented software is open source (GPL 3.0 license).
Requirements
- Cytoscape 2.6.3 or later (http://www.cytoscape.org/)
- Java 1.6
- Tested on Cytoscape 2.6.3 and 2.7.0 on Windows, Linux and Mac OS X.
Data
The data files for the evolutionary analysis are provided below. The format of the data is described in the manual.
Additional data sources
While it requires some effort, there exist other sources of data which can be integrated to provide the input for the evolutionary analysis using our plugins. Protein trees can be obtained using tree reconstruction methods or downloaded from databases such as TreeFam. Weighted protein-protein interaction networks are provided for example by STRING. STRING also provides a high-level network induced by orthologous clusters which is similar to our ancestral (root) network. Also many network alignment techniques (e.g. NetworkBlast, Graemlin) provide a representation of a conserved network (e.g. alignment graph) which can be interpreted and visualized similarly to our ancestral network.